Home : HD-Peptide Microarray synthesis
High-density peptide microarrays
We have developed a complete set of tools for design, synthesis and recording of high-density peptide microarrays [1-3]. The arrays can contain up to 2 million predefined peptide sequences and they are well suited for identification of antibody peptide epitopes and for mapping of peptide recognition motifs for other proteins (receptor ligands, protein kinases etc.). Work on the high-density peptide microarrays was initiated in 2003 in the EU FP6 programme “Vaccines_To_Genomes“ and was continued in the EU FP7 programme “PepChipOmics” launched in 2008. During this program, high-density peptide arrays each with more than 500.000 individually defined peptide sequences were synthesized and tested in epitope screening assays. In the EU FP7 programme “HIPAD“ the technique has been further refined and used in generation of binding data for polyclonal antibodies and other proteins with affinity for linear peptides.
Each array is generated within a small (ca. 1 x 2 cm) rectangular area on a functionalized microscope slide. The individual peptide fields are square-shaped with a freely definable side length from 10 x 10 µm and up to 1000 x 1000 µm. When the smallest field size is used, more than 2 million peptide fields can – in principle – be made in each array. In most cases it is advisable to work with larger fields, e.g. 20 x 20 µm fields to facilitate recording of the arrays after binding of ligands. With this field size you still have more than 500.000 peptide fields in each array and at this resolution most array scanners are able to record the individual peptide fields.

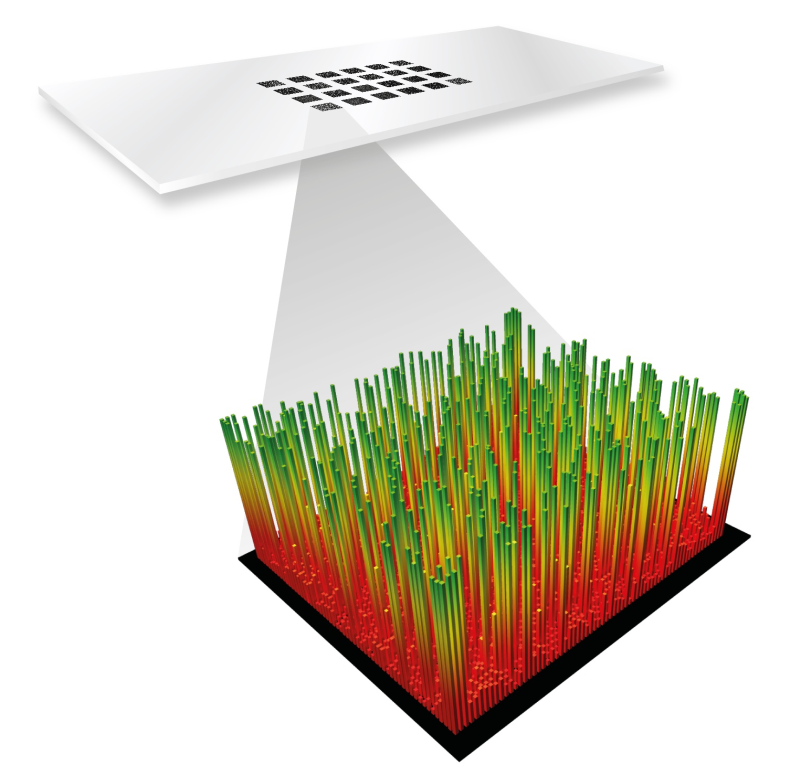
A single high density peptide microarray can contain complete sets of overlapping peptides from hundreds of average sized proteins – or it can be designed to contain multiple variants of all peptides that can be derived from one or a few proteins.
Contact us for a communication about your needs and thoughts related to high density peptide microarrays. Based on your FASTA sequences or lists of individual peptide sequences we can make the array layouts and synthesize the arrays. As an additional service, we can incubate the arrays with your ligands and record the results for further processing in your preferred data format.
Typical turnover time is about 2 weeks after receipt of your sequences and reagents for probing of the arrays.
Users that consume HD-peptide arrays in larger numbers can get free access to our design- and analysis software together with online instructions and training in use of the programs.
[1]: Mol Cell Proteomics (2012), PMID 22984286
[2]: PloS One (2013), PMID:23894373
[3]: Mol Cell Proteomics (2015), PMID:25922409
[4]: Scientific Reports 6, 22030 (2016); doi: 10.1038/srep22030